 |
This workshop is partially supported
Overlapping Cervical Cytology Image Segmentation Challenge - ISBI 2014
[May 1, 2014] Challenge Ranking Results Released! See details.
[Mar 16, 2014] Submission of result is accepted! See details.
[Feb 23, 2014] Synthetic Cytology Images Generator Code Released! See Description page for details.
[Feb 05, 2014] 810 Test synthetic images released! See Dataset page for details.
[Jan 11, 2014] Training dataset and evaluation code released! See Dataset page.
[Dec 26, 2013] Announcement! All participants need to register as an non-author to ISBI 2014. [link]
[Dec 16, 2013] Challenge website launched!
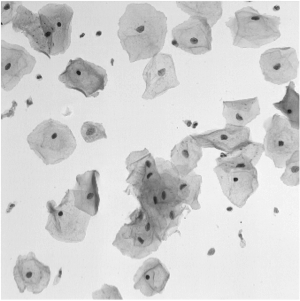 |
The automated detection and segmentation of overlapping cells using microscopic images obtained from Pap smear can be considered to be one of the major hurdles for a robust automatic analysis of cervical cells. The Pap smear is a screening test used to detect pre-cancerous and cancerous processes, which consists of a sample of cells collected from the cervix that are smeared onto a glass slide and further examined under a microscope (see the left figure). The main factors affecting the sensitivity of the Pap smear test are the number of cells sampled, the overlap among these cells, the poor contrast of the cell cytoplasm, and the presence of mucus, blood and inflammatory cells. Automated slide analysis techniques attempt to improve both sensitivity and specificity by automatically detecting, segmenting and classifying the cells present on a slide. In this challenge, the targets are to extract the boundaries of individual cytoplasm and nucleus from overlapping cervical cytology images. |
The First Segmentation of Overlapping Cervical Cells from Extended Depth of Field Cytology Image Challenge is held under the auspices of the IEEE International Symposium on Biomedical Imaging (ISBI 2014) held in Beijing, China on April 28th - May 2nd, 2013.