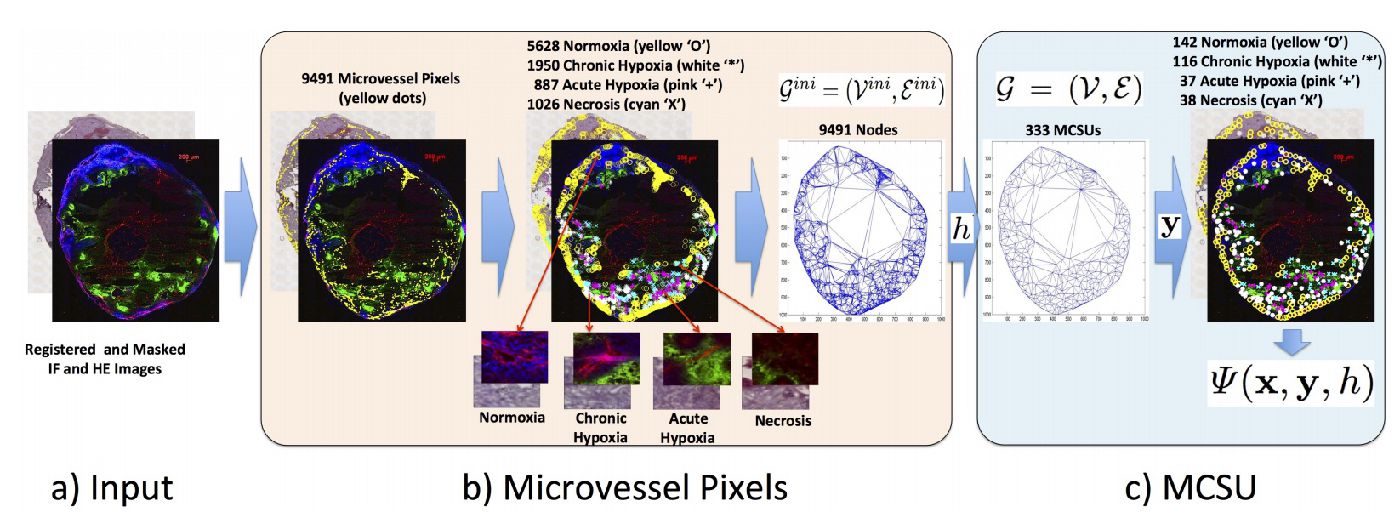
Flexible and Latent Structured Output Learning: Application to Histology
Baseline for the structured output learning approach estimating the amount of hypoxic regions in microscopy imaging of tumor tissues.
Compare the use of structure output prediction models with a new deep learning model for estimating the amount of hypoxic regions in microscopy imaging of tumor tissues.

Baseline for the structured output learning approach estimating the amount of hypoxic regions in microscopy imaging of tumor tissues.
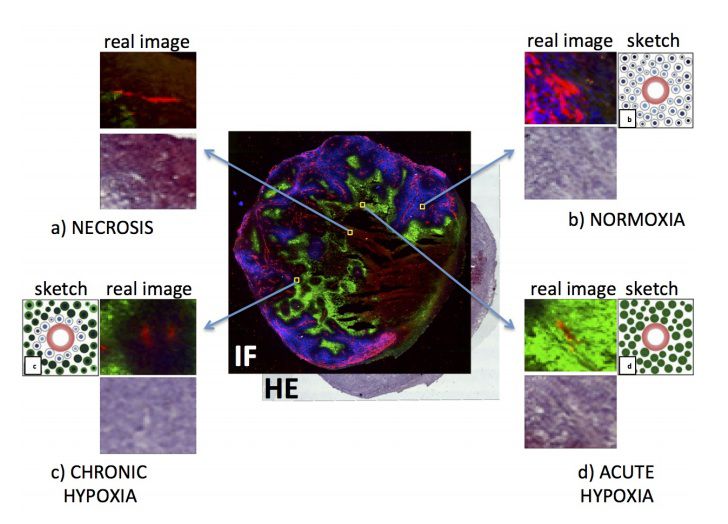
Classifier of local image regions into normoxia or hypoxia.
There are two datasets available:
1. Cross validation sets for the training/testing of the microvessel pixel classification
2. Cross validation sets for the training/testing of the flexible and latent structured output model.
They are available as two mat files, and we also provide an m-file to read those sets. The structure of the datasets is self-explanatory, but do not hesitate to email Gustavo Carneiro in case you have questions.
If you use this dataset, please cite the following work:
Gustavo Carneiro, Tingying Peng, Christine Bayer and Nassir Navab. Weakly-supervised Structured Output Learning with Flexible and Latent Graphs using High-order Loss Functions. In Proceedings of the International Conference on Computer Vision (ICCV) 2015.